Metabolic interactions: gut microbiome and host-pathogen interactions
One of the next frontiers of microbial research are metabolic interactions between species. In several collaborative projects we investigate fundamental questions on which metabolites are exchanged between microbes and with the host, and how their own metabolism operates in such consortia.
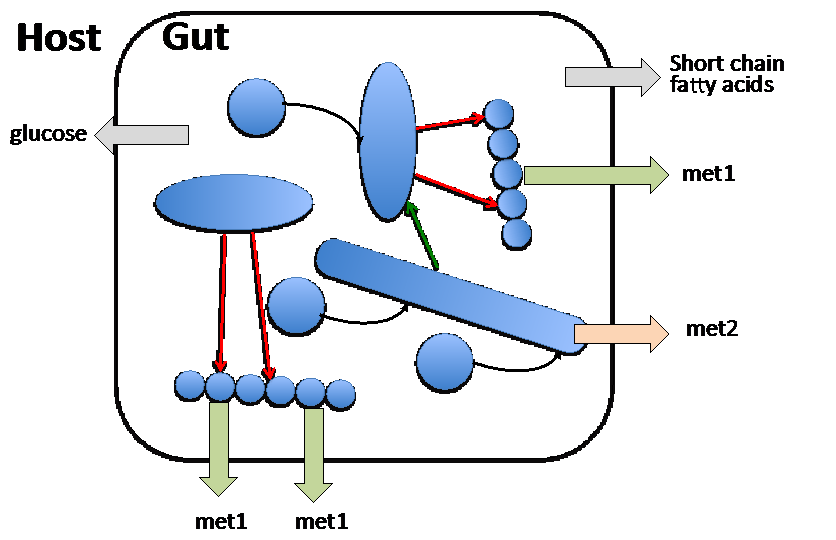
Ourmain active project in this area concerns gut microbial communities - specifically, a recently established model community consisting of 12 species in the mouse gut (OMM12). Metabolic interactions are very important in the gut, where complex communities often confer benefits to their host. However, these interactions are difficult to decipher and the otherwise successful sequencing projects provide only very limited mechanistic insights. Over the last years, our lab developed concepts and experimental methods to unravel metabolic or signaling interactions within communities through in vitro cultivation experiments, high-throughput metabolomics and isotopic labeling experiments that allow us to generate hypotheses on species interactions. This project is part of a long-standing collaboration with the external pageMacpherson labcall_made at the Inselspital in Bern that hosts a gnotobiological mouse facility for in vivo experiments.
In a past project, we focused on the pathogen Mycobacterium tuberculosis in the context of the external pageTbXcall_made project and on natural consortia in food biotechnology. A huge challenge are the complex metabolic interactions in the human gut microbiome (about 1 kg of bacteria) that influence our well being in many ways. In close collaboration with the external pageMacpherson labcall_made at the Inselspital in Bern, we investigated this metabolic handshake between the gut microbiome and gnotobiological mice as well as humans. To this end we employe dynamic stable tracer labeling experiments and a high-throughput metabolomics platform that allows us to detect about 500 metabolites in a given sample per minute. Thus, we go beyond simply counting who is there but venture into what are they doing by focusing on mechanisms and causality.
References
- Meier K, Trouillon J, Li H, Lang M, Fuhrer T, Zamboni N, Sunagawa S, Macpherson A & Sauer U (2023). Metabolic landscape of the male mouse gut identifies different niches determined by microbial activities. Nat Metab 5, 968–980. external pagehttps://doi.org/10.1038/s42255-023-00802-1call_made
- Macpherson A & Sauer U (2023). Secrets of microbiota drug metabolism. Nat Med, 29, 537-538. external pagehttps://doi.org/10.1038/s41591-023-02227-5call_made
- Pérez-Escriva P, Fuhrer T & Sauer U (2022). Distinct N and C Cross-Feeding Networks in a Synthetic Mouse Gut Consortium. mSystems. external pagehttps://doi.org/10.1128/msystems.01484-21call_made
- Machova I, Snasel J, Zimmermann M, Laibitz D, Plocinski P, Oehlmann W, Singh M, Dostal J, Sauer U, Pichova I (2014). Mycobacterium tuberculosis phosphoenolpyruvate carboxykinase is regulated by redox mechanisms and interaction with thioredoxin. J Biol Chem. 289:13066-78. external pagePubmedcall_made
- Zimmermann M, Thormann V, Sauer U & Zamboni N (2013). Non-targeted profiling of coenzyme A thioesters in biological samples by tandem mass spectrometry. Anal Chem. 85:8284-90.
- Watanabe S, Zimmermann M, Goodwin MB, Sauer U, Barry CE 3rd, Boshoff HI (2011). Fumarate reductase activity maintains an energized membrane in anaerobic M. tuberculosis. PLoS Pathogens. 7:e1002287.
- Rühl M, Hardt WD, Sauer U (2011). Revealing subpopulation specific metabolic pathway usage in mixed cultures by reporter protein-based 13C analysis. Appl Environ Microbiol. 77(5):1816-21. external pagePubmedcall_made
