Research
The overarching theme of the lab is understanding the coordination of cellular networks, metabolism and regulation in particular. Development of high-throughput technology and conceptually new approaches place us in a unique position.
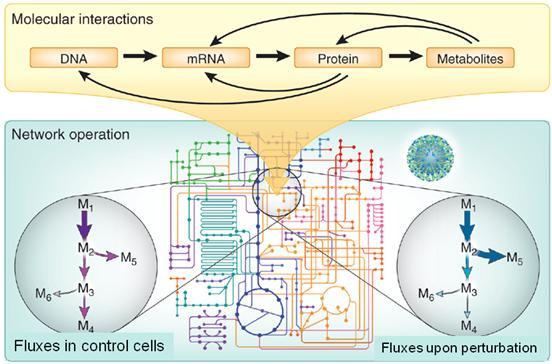
Overlapping and intertwined regulatory networks of genes, mRNAs, proteins and metabolites enable adaptation to ever changing environments through multiple molecular mechanisms. At best we have a vague understanding which of these mechanisms really matter for changing particular cellular functions. We quantitatively identify the key regulation mechanisms that control metabolism in bacteria and yeast (see Figure) and how metabolism feeds back information to the regulation systems or is the signal generator in the first place. Currently we focus on the short-lived regulatory metabolite-protein interaction and the functional relevance of protein phosphorylation, two presently underdeveloped areas. Beyond model microbes, we work on basic metabolic questions with pathogenic Mycobacteria and interacting microbial consortia such as the gut microbiome.
The competitive team advantage of our biologists, engineers, and computer scientists are high-throughput and dynamic experimental technologies for metabolomics and 13C-based flux analysis. Different types of mathematical models and computational analyses are then used to quantitatively relate metabolomics, expression and proteomics data to the functional network output in terms of fluxes.
Further details can be found in the subpages.
References
- Gerosa L, Haverkorn van Rijsewijk B. R. B, Christodoulou D, Kochanowski K, Schmidt T. S. B, Noor E, & Sauer U (2015). Pseudo-transition Analysis Identifies the Key Regulators of Dynamic Metabolic Adaptations from Steady-State Data. Cell Syst., vol. 1, no. 4, pp. 270–282, Oct. 2015.
- Link H, Fuhrer T, Gerosa L, Zamboni N, and Sauer U (2015). Real-time metabolome profiling of the metabolic switch between starvation and growth. Nat. Methods, vol. 12, no. 11, pp. 1091–1097
- Chubukov V, Gerosa L, Kochanowski K & Sauer U (2014). Coordination of microbial metabolism. Nature Rev Microbiol. 12:327-40.
- Link H, Kochanowski K & Sauer U (2013). Systematic identification of allosteric protein-metabolite interactions that control enzyme activity in vivo. Nature Biotechnol. 31: 357-61.
- Kochanowski K, Sauer U & Chubukov V (2013). Somewhat in control – the role of transcription in regulating microbial metabolic fluxes. Curr Opin Biotechnol. 24: 987-933.
- Gerosa L, Kochanowski K, Heinemann M & Sauer U (2013). Dissecting specific and global transcriptional regulation of bacterial gene expression. Mol Sys Biol. 9: 658.
- Büscher JM, Liebermeister W, Jules M, Uhr M, Muntel J – 31 others – Bessieres P, Devine K, Harwood CR, Hecker M. Jarmer H, Klipp E, Lewis P, Molina F, Noirot P, Schwikowski B, van Dijl JM, Wilkinson AJ, Stelling J, Aymerich S & Sauer U (2012). Global network reorganization during dynamic adaptations of B. subtilis metabolism. Science 335: 1099-1103.
- Heinemann M & Sauer U (2010). Systems biology of metabolism. Curr Opin Microbiol. 13: 337-343.
