Escherichia coli metabolism
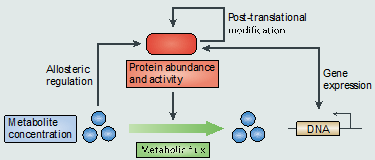
Ultimately we want to quantitatively and mechanistically understanding (and predict) how genes, proteins, and metabolites interact to enable cells to thrive under many different and changing conditions – and to engineer such networks effectively for biotech applications. For E. coli metabolism and its regulation we bring all our methods and approaches to bear on a single focus. Starting from considering gene expression, phosphorylation and enzyme kinetics separately, we believe the timing to be right to work towards dynamic mechanistic models that capture the key processes across time scales from seconds to hours of, for example, nutritional transitions. Additionally several genome-scale studies are ongoing that include metabolomics of nearly 4000 single gene deletions, functional genomics of the entire collection of >1000 genes with unknown function, and functional analysis of protein phosphorylation. In several projects we collaborate with the computational biology labs of external pageDennis Vitkupcall_made at Columbia, Jörg Stelling at ETH, and external pageTerry Hwacall_made a UCSD.
References
- Gerosa L, Haverkorn van Rijsewijk B. R. B, Christodoulou D, Kochanowski K, Schmidt T. S. B, Noor E, & Sauer U (2015). Pseudo-transition Analysis Identifies the Key Regulators of Dynamic Metabolic Adaptations from Steady-State Data. Cell Syst., vol. 1, no. 4, pp. 270–282, Oct. 2015.
- Chubukov V, Gerosa L, Kochanowski K & Sauer U (2014). Coordination of microbial metabolism. Nature Rev Microbiol. 12:327-40.
- Gerosa L, Kochanowski K, Heinemann M & Sauer U (2013). Dissecting specific and global transcriptional regulation of bacterial gene expression. Mol Sys Biol. 9: 658.
- Link H, Kochanowski K & Sauer U (2013). Systematic identification of allosteric protein-metabolite interactions that control enzyme activity in vivo. Nature Biotechnol. 31: 357-61.
- Schütz R, Zamboni N, Zampieri M, Heinemann M & Sauer U (2012). Multidimensional optimality of microbial metabolism. Science 336: 601-604.
