Quantitative metabolomics
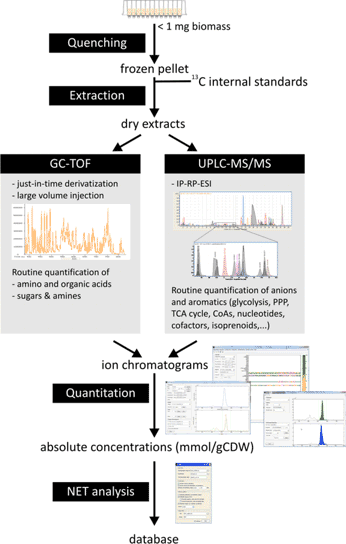
Whenever accurate measurements of metabolite pools are necessary, we rely on targeted metabolomics by either LC-MS/MS, GC-TOF, or GC-MS depending on the analytes. The standard workflow for quantification of key intermediates of central carbon metabolism relies on ion pairing LC-MS/MS. This method allows to separate most isomers groups in central metabolism and yet offers a broad coverage of heterogeneous classes of compounds (i.e. organic acids, amino acids, sugar phosphates, nucleotides, CoA derivatives, etc). In parallel, we use various specialized methods for the analysis of special compounds which are hardly captured with general approaches.
To ensure the highest standards of quality, we systematically employ isomeric 13C labeled internal standards for all analytes of interest. Precise and rapid quantification of hundreds of metabolites in hundreds of samples is ensured by own software that employs machine learning to robustly process real-life chromatographic traces without user supervision. Finally, quality of measurements is assessed by special software that checks for thermodynamic consistency of the whole data set.
- Zimmermann M, Sauer U, Zamboni N, Quantification and Mass Isotopomer Profiling of α-Keto Acids in Central Carbon Metabolism, Anal Chem 2014
- Zimmermann M, Thormann V, Sauer U, Zamboni N, Non-targeted profiling of coenzyme A thioesters in biological samples by tandem mass spectrometry, Anal Chem 2013; 85 (17): 8284–8290
- Büscher J, Moço S, Sauer U, Zamboni N, Ultra-high performance liquid chromatography-tandem mass spectrometry method for fast and robust quantification of anionic and aromatic metabolites, Anal Chem 2010; 82(11):4403-4412
- Ewald JC, Heux S, Zamboni N, High-throughput quantitative metabolomics: a workflow for cultivation, quenching and analysis of yeast in a multi-well format, Anal Chem 2009; 81(9):3623-3629
- Büscher JM, Czernik D, Ewald JC, Sauer U, Zamboni N, Cross-Platform Comparison of Methods for Quantitative Metabolomics of Primary Metabolism, Anal Chem 2009; 81(6):2135-2143
- Zamboni N, Kümmel A, Heinemann M, anNET: a tool for network-embedded thermodynamic analysis of quantitative metabolome data, BMC Bioinformatics 2008, 9:199
- Sévin DC, Kuehne A, Zamboni N, Sauer U. Biological insights through nontargeted metabolomics, Curr Opin Biotechnol. 2015:34 external pagedoicall_made
- Zamboni N, Sauer U, Novel biological insights through metabolomics and 13C-flux analysis, Curr Opin Microbiol 2009; 12(5):553-8
