Research
Motivation
Cellular processes in health and disease emerge from dynamic molecular networks of enormous complexity. Proteins represent the most abundant class of cellular network components. More than 10 billion molecules of over 100000 different proteins are estimated to form into stable and transient complexes that are organized into dynamic regulatory networks that coordinate the various processes in a typical human cell. This seeming molecular complexity represents an enormous analytical challenge. Not surprisingly only a small fraction of the protein networks in human cells has been characterized to date. In our group, we develop and apply new quantitative mass spectrometry approaches to systematically study the topology and temporal control of protein networks at high through put and quantitative accuracy.
We are specifically interested in dynamic networks controlling cell signaling and epigenetic gene regulation linked to human diseases. We believe that understanding the topology and temporal organization of such networks represents an important conceptual framework to better interpret and predict cellular phenotypes emerging from mutated patients’ genomes and in response to drug action.
Approach
We develop and apply various approaches for the biochemical enrichment of protein complexes (size exclusion chromatography, affinity purification and biotin proximity labeling). These techniques are combined with mass spectrometry based proteomics techniques to reveal composition, stoichiometry and temporal control of protein complexes. Computational tools for visualization of proteomic data and their integration with genomic and phenomic data are used to infer new hypothesis that can be tested by specific functional experiments.
Specific projects of the group
Topology and dynamics of cell signaling networks
- Characterization the endothelin signalling system and its role in cell growth (Alex Schäfer in collaboration with ACTELION)
- The protein interaction landscape of the human kinome in health and disease (Audrey van Drogen, Marija Bujan, Anton Vichalkovsky, Rodolfo Ciuffa, Martin Mehnert)
- Signaling dynamics controling the activation of primary T cells (Simon Hauri, Etienne Caron in collaboration with Romain Roncagalli and Bernard Malissen)
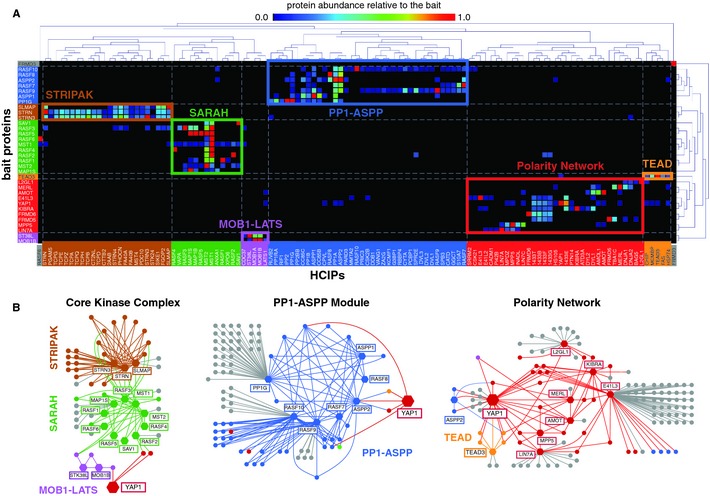
- Topology and temporal organization of signaling complexes controlling tissue homeostasis by the human HIPPO pathway (Federico Uliana)
Functional proteomics of epigenetic gene regulation
- Modulation of human histone methyl transferase complexes in human cancer (Fabian Frommelt)
- Changes in chromosomal protein complexes associated with X-chromosome inactivation (Andrea Fossati in collaboration with the lab of Anton Wutz, Molecular Health Sciences, ETH)
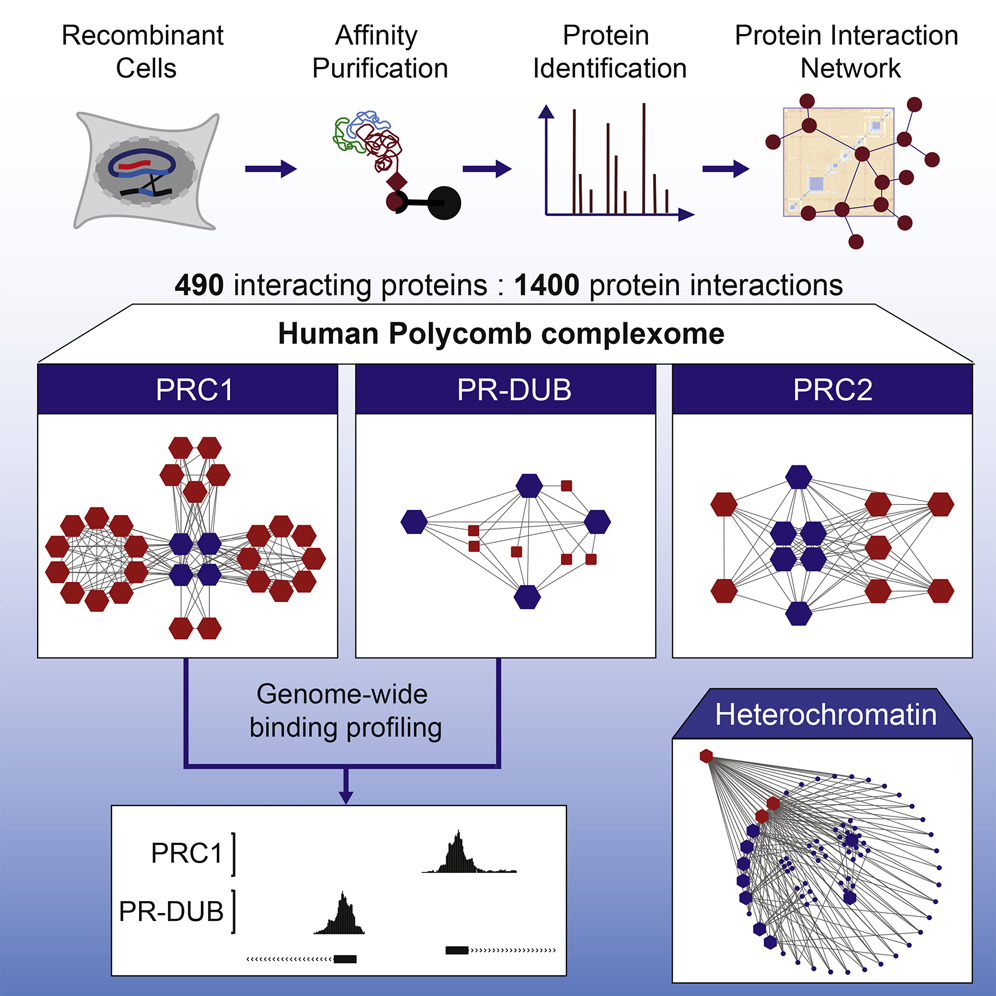
- The molecular interaction landscape of human Polycomb complexes (Simon Hauri in collaboration with Christian Beisel, BSSE Basel)